Filtering Raw Patterns in HDF5¶
Process raw EBSD images in HDF5 file and save the Kikuchi signal.
[22]:
import os
data_dir = "../../../xcdskd_example_data/GaN_Dislocations_1"
h5FileNameFull=os.path.abspath(data_dir+"/hdf5/GaN_Dislocations_1.hdf5")
#print(h5FileNameFull)
nbin=1 # binning factor for pattern processing
[23]:
%load_ext autoreload
%autoreload 2
%matplotlib inline
The autoreload extension is already loaded. To reload it, use:
%reload_ext autoreload
[24]:
import numpy as np
import skimage.io
import time
import sys
import h5py
from shutil import copyfile
from aloe.image import kikufilter
from aloe.plots import plot_image
[25]:
# close HDF5 file if still open
if 'f' in locals():
f.close()
# split off the extension
h5FileName, h5FileExt = os.path.splitext(h5FileNameFull)
h5FilePath, h5File =os.path.split(h5FileNameFull)
# copy to new file
src = h5FileName+h5FileExt
dst = h5FileName+'_processed'+h5FileExt
copyfile(src, dst)
# use new filename from here to work on
h5FileName = h5FileName+'_processed'
f=h5py.File(h5FileName+h5FileExt, "a") # append mode
#f=h5py.File(h5FileName+h5FileExt, "r")
#print('HDF5 full file name: '+h5FileNameFull)
#print('HDF5 File: '+h5FileName+h5FileExt)
#print('HDF5 Path: '+h5FilePath)
[26]:
DataGroup="/Scan/EBSD/Data/"
HeaderGroup="/Scan/EBSD/Header/"
Patterns = f[DataGroup+"RawPatterns"]
XIndex = f[DataGroup+"X BEAM"]
YIndex = f[DataGroup+"Y BEAM"]
MapWidth = f[HeaderGroup+"NCOLS"].value
MapHeight= f[HeaderGroup+"NROWS"].value
PatternHeight=f[HeaderGroup+"PatternHeight"].value
PatternWidth =f[HeaderGroup+"PatternWidth"].value
print('Pattern Height: ', PatternHeight)
print('Pattern Width : ', PatternWidth)
PatternAspect=float(PatternWidth)/float(PatternHeight)
print('Pattern Aspect: '+str(PatternAspect))
print('Map Height: ', MapHeight)
print('Map Width : ', MapWidth)
Pattern Height: 256
Pattern Width : 336
Pattern Aspect: 1.3125
Map Height: 50
Map Width : 52
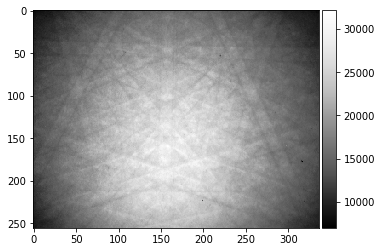
[27]:
RawPattern=Patterns[1000,:,:]
plot_image(RawPattern)
#skimage.io.imsave('raw_pattern.tiff', RawPattern, plugin='tifffile')
[28]:
# TRY OPTIONAL: make static background from average of all patterns in map
#StaticBackground=np.mean(Patterns[:], axis=0)
#f[HeaderGroup+"StaticBackground"] = StaticBackground
[29]:
StaticBackground=f[DataGroup+"StaticBackground"]
plot_image(StaticBackground)
#np.savetxt('StaticBackground.txt',StaticBackground )

[30]:
PatternFlat=RawPattern/StaticBackground
plot_image(PatternFlat)
#from scipy import misc
#misc.imsave('Pattern_Flatfielded.png',RawPattern)
#scipy.misc.toimage(image_array, cmin=0.0, cmax=...).save('outfile.jpg')
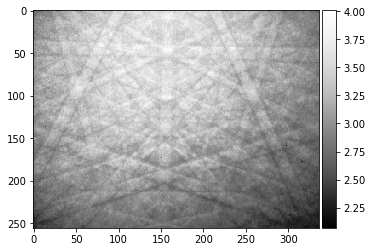
We test the parameters for the processing of the raw patterns in the HDF5 file:
[31]:
kpattern=kikufilter.process_ebsp(raw_pattern=RawPattern,
static_background=StaticBackground, sigma=20,
clow=1.0, chigh=99.0, dtype=np.uint8, binning=nbin)
plot_image(kpattern)
print(kpattern.dtype)

uint8
We also check the static background:
[32]:
bgpattern=kikufilter.process_ebsp(raw_pattern=StaticBackground,
static_background=None, sigma=15,
clow=0.5, chigh=99.5, dtype=np.uint16, binning=nbin)
plot_image(StaticBackground)
plot_image(bgpattern)
print(bgpattern.dtype)

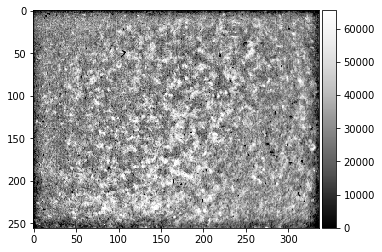
uint16
Save Processed Patterns in HDF5¶
[33]:
nPatterns=MapWidth*MapHeight
print('Number of patterns: ',nPatterns)
# store *processed* patterns in HDF5
# create empty data set, compress (max=9) using hdf5-gzip filter
dataset_name=DataGroup+'Patterns'
if not (dataset_name in f):
print('create: ', dataset_name)
dset_processed=f.create_dataset(dataset_name,
(nPatterns,PatternHeight//nbin,PatternWidth//nbin),
dtype=np.uint8,
chunks=(1,PatternHeight//nbin,PatternWidth//nbin))
#compression='gzip',compression_opts=9)
else:
# overwrite existing data
print('data set already existing: overwriting ', dataset_name)
dset_processed=f[dataset_name]
#nPatterns=5
# this is very slow...
for p in range(nPatterns):
img_raw = np.copy(Patterns[p])
img = kikufilter.process_ebsp(raw_pattern=img_raw,
static_background=StaticBackground, sigma=15,
clow=1, chigh=99, dtype=np.uint8, binning=nbin)
# update HDF5 data set
dset_processed[p]=img
# live update progress info
if ( p % 10 == 0 ):
progress=100.0*(p+1)/nPatterns
sys.stdout.write("\rtotal patterns: %5i current:%5i progress: %4.2f%%"
% (nPatterns, p, progress) )
sys.stdout.flush()
f.close()
Number of patterns: 2600
create: /Scan/EBSD/Data/Patterns
total patterns: 2600 current: 2590 progress: 99.65%
[34]:
f.close()
Test: Plotting the processed patterns¶
[35]:
f=h5py.File(h5FileName+h5FileExt, "r")
[36]:
kPatterns = f[DataGroup+"Patterns"]
plot_image(kPatterns[2500])

[37]:
indexmap = f[HeaderGroup+"indexmap"]
def get_npa(x, y, patterns, indexmap, nn=0):
""" get neighbor pattern average
use -nn..+nn neighbors,
i.e. at nn=1, there will be 8 neighbors = 9 pattern average
patterns in assumed to be 1D array of 2D patterns
index of pattern as function of x,y is in indexmap
"""
ref_pattern = np.copy(patterns[indexmap[x,y]]).astype(np.int64)
npa = np.zeros_like(ref_pattern, dtype=np.int64)
for ix in range(x-nn,x+nn+1):
for iy in range(y-nn, y+nn+1):
print(ix, iy)
if ((ix<0) or (ix>=patterns.shape[1]) or (iy<0) or (iy>=patterns.shape[0])):
npa = npa + ref_pattern
else:
npa = npa + patterns[indexmap[ix,iy]]
return npa
[38]:
img_npa = get_npa(24, 24, kPatterns, indexmap, nn=1)
plot_image(img_npa)
23 23
23 24
23 25
24 23
24 24
24 25
25 23
25 24
25 25

[ ]: